MicFly: Rapid, Single-Cell Phenotyping and Absolute Quantification of Bacterial Microbiota
Researchers at Pitt have developed a new method that is a significant improvement over existing techniques to analyze the microbiota in the intestine. This method is ready to license or partner for further developing applications in translational research and clinical diagnostic. This technology combines fluorescent labels and Hybridization Chain Reaction (HCR) with flow cytometry to rapidly identify and quantify two or more characteristics of a bacterium in a single sample at the single-cell level. This approach represents a significant advancement over traditional sequencing methods by offering a fast, quantitative, and accurate measurement of bacterial identity and crucial surface characteristics, such as host-antibody binding, in complex samples like the intestinal microbiota.Microbiota research is primarily sequencing-based, subjected to PCR amplification bias, and restricted to relative abundances. Having a granular understanding of the microbiota for clinical and diagnostic purposes is hampered by the lack of single cell methods. To this end, Pitt researchers have developed Microbiome Flow Cytometry (MicFly), a single-cell technology combining spectral flow, RNA hybridization chain reaction, and surface protein-targeted antibodies.
Description
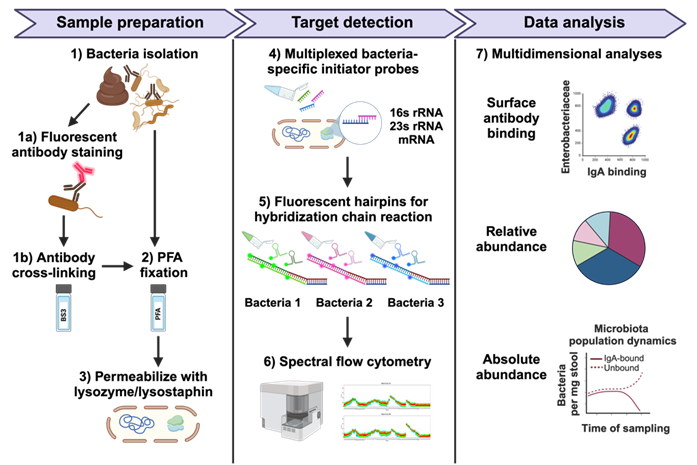
The MicFly technology couples the measurement of a bacterium's surface phenotype with its genomic identity. First, the bacterium in a sample is stained with a fluorescently labeled ligand that binds to a surface target, such as a host antibody (e.g., IgA, IgM, or IgG). Crucially, this labeled complex is chemically cross-linked to the bacterium's surface and the cell is fixed and permeabilized, a unique step that retains the surface-bound antibodies throughout the entire protocol for accurate phenotyping. The innovative aspect of the method lies in the next step: Hybridization Chain Reaction (HCR). HCR is performed inside the permeabilized bacterium to generate a second fluorescent signal that is specific to a target bacterial DNA or RNA sequence (e.g., phylum, genus, or species) . The dual-labeled bacterium is then analyzed using fluorescence-activated sorting (Flow Cytometry). This single assay run simultaneously yields both the surface binding characteristic and the bacterial identity for thousands of individual cells, enabling absolute and relative quantification of complex bacterial populationsApplications
- Gastrointestinal and Infectious Disease Diagnostics: Rapid and quantitative identification of pathogens, such as Clostridium difficile, and simultaneous measurement of their virulence factor expression in patient samples.- Microbiome Research and Therapeutic Development: Efficiently characterizing the structure and dynamics of complex microbiota for studies related to diseases like Inflammatory Bowel Disease (IBD) or Necrotizing Enterocolitis.
- Mucosal Immunity and Host-Microbe Interaction Studies: Analyzing bacteria bound by host antibodies (IgA, IgM, IgG) in samples like stool to provide insight into mucosal immune responses and monitor early life development .
- Pre-Clinical and In Vivo Model Analysis: Quantifying and tracking the members of defined microbial consortia in gnotobiotic animal models with precision comparable to qPCR.
- Pharmaceutical and Probiotic Quality Control: Absolute quantification of specific microbial members in defined consortia or fermentation products.
Advantages
- Rapid, Single-Cell Analysis: Provides faster, more accurate identification and quantification of bacteria at the single-cell level compared to conventional sequencing methods, addressing the critical need for speed in clinical use.- Dual-Characteristic Measurement: Simultaneously determines a bacterium's identity (via HCR) and a crucial surface characteristic (e.g., binding of host antibodies like IgA) in a single assay run.
- Absolute Quantification: Accurately determines both the absolute and relative abundance of different bacterial populations within a complex sample, a significant advantage for tracking true population changes.
- Preserved Binding Information: Employs a unique cross-linking step to retain surface-bound ligands on the bacteria throughout the protocol, which is essential for stable and accurate antibody-phenotyping.
- Direct Gene Expression Measurement: Capable of measuring bacterial gene expression (e.g., toxin production or mRNA levels) at the single-cell level concurrent with species identification.
- Allows for the targeted RNAsequencing of individual bacterial taxa within a mixed population.
Invention Readiness
The technology is an advanced prototype that has been successfully demonstrated in a highly relevant, near-operational environment. Key data generated includes the absolute and relative quantification of diverse bacterial populations, simultaneous measurement of bacterial identity and host antibody binding (e.g., IgA-bound bacteria), and direct measurement of bacterial gene expression in a single run of the assay. This robust validation has been performed across complex biological matrices, including human patient samples (stool and infected tissue) and gnotobiotic mouse models. Future efforts will focus on optimizing the assay for high-throughput commercial and clinical laboratory use and expanding the panel of bacterial targets for broad-spectrum infectious disease and microbiome applications.IP Status
https://patents.google.com/patent/WO2025076345A1Related Publication(s)
Christine M. Tin, Bianca Cordazzo Vargas, Darryl A. Abbott, Aditi R. Lohar, Tiffany C. Taylor, Ivan A. Valishev, Sarah Weinshel, Vung Lian, Kacey J. Sullinger, Matthew Butoryak, Michael A. Silverman, Liat Shenhav, William H. DePas, Timothy W. Hand. Single-cell quantification of the microbiota by flow cytometry: MicFLY.
bioRxiv 2025.09.18.677130; doi: https://doi.org/10.1101/2025.09.18.677130
Johnson-Hence, C. B., Gopalakrishna, K. P., Bodkin, D., Coffey, K. E., Burr, A. H. P., Rahman, S., Rai, A. T., Abbott, D. A., Sosa, Y. A., Tometich, J. T., Das, J., & Hand, T. W. (2023). Stability and heterogeneity in the antimicrobiota reactivity of human milk-derived immunoglobulin A. Journal of Experimental Medicine, 220(8). https://doi.org/10.1084/jem.20220839
Burr, A. H. P., Ji, J., Ozler, K., Mentrup, H. L., Eskiocak, O., Yueh, B., Cumberland, R., Menk, A. V., Rittenhouse, N., Marshall, C. W., Chiaranunt, P., Zhang, X., Mullinax, L., Overacre-Delgoffe, A., Cooper, V. S., Poholek, A. C., Delgoffe, G. M., Mollen, K. P., Beyaz, S., & Hand, T. W. (2023). Excess Dietary Sugar Alters Colonocyte Metabolism and Impairs the Proliferative Response to Damage. Cellular and Molecular Gastroenterology and Hepatology, 16(2), 287–316. https://doi.org/10.1016/j.jcmgh.2023.05.001
